Sequence-Based Macromolecules
The module for sequence-based macromolecules allows the definition and storage of large molecules based on repeating building blocks. Currently, proteins are supported, with other macromolecules such as RNA and DNA planned for the future.
(1) SBMMs cannot yet be used as components of chemical reactions; this feature is scheduled for a future release. (2) Some calculations in the SMBMM model are not yet supported.
SBMM concept and SBMM samples
SBMMs are implemented as elements in the ELN. That means that they can be retrieved from the elements' list and have a detailed view upon clicking on the single entries in the elements list. SBMMs consist of the properties, identifiers and characteristics of the SBMM concept (basically the molecule) and the SBMM sample. The SBMM concept is identified by the sequence of the SMBB. The SBMM sample is characterized by additional properties that are batch specific. The SBMM concept can have multiple samples (each a physical representation of the SBMM concept). While the SBMM concept exists just once in the database of an ELN instance (and can be used by differnt users) the samples assigend to an SBMM concept belong to a user and can only by seen and edited by the user.
Creation of an SBMM concept
In a first step, the type of sequence-based macromolecule is chosen. At the moment, only the type "protein" is supported. The user can either (1) re-use exising proteins that are already stored of the ELN database and can be retrieved from the elements' list (use drag and drop field) (2) search a protein in the ELN database or in uniprot and create a new sample assigned to it (dropdown options) (3) create a protein from scratch (dropdown options)
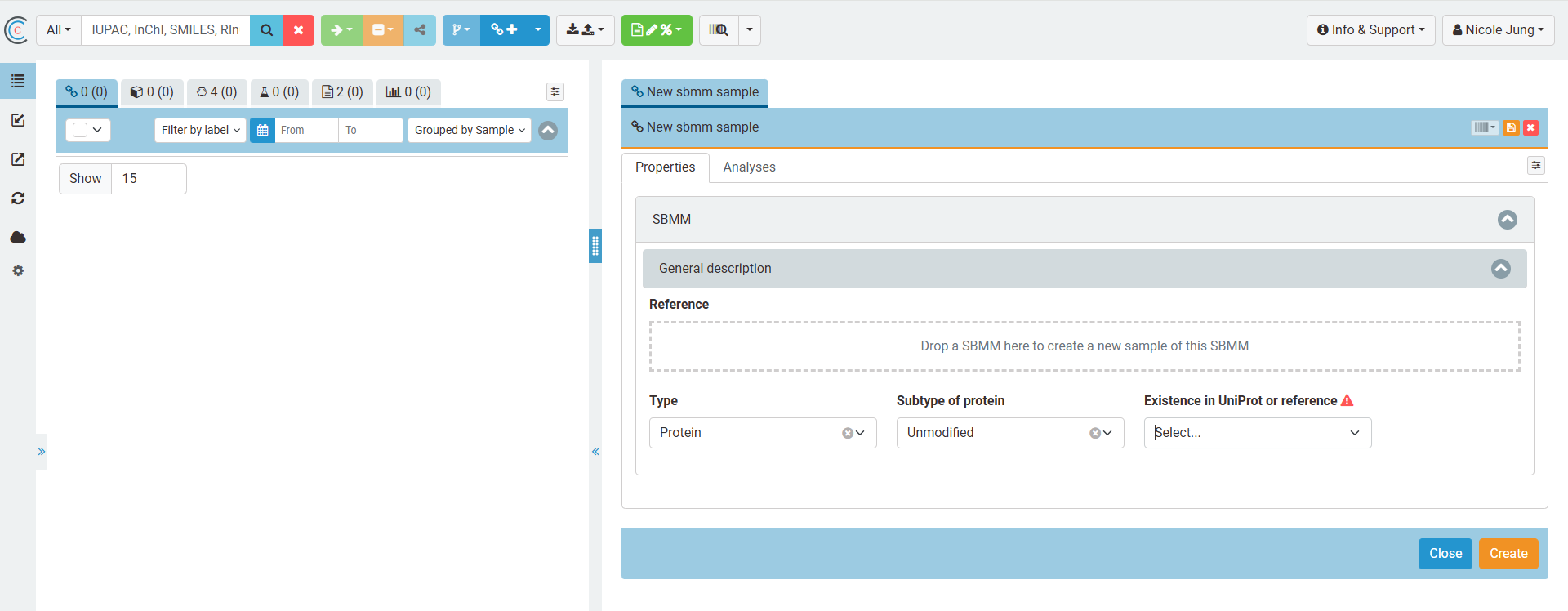
Depending on how the protein is gained or where data describing the protein can be gained from, the user has the option to define SBMMs for that the characteristics correspond to UniProt entries, that are derived from Uniprot and modified, and those that are not known.
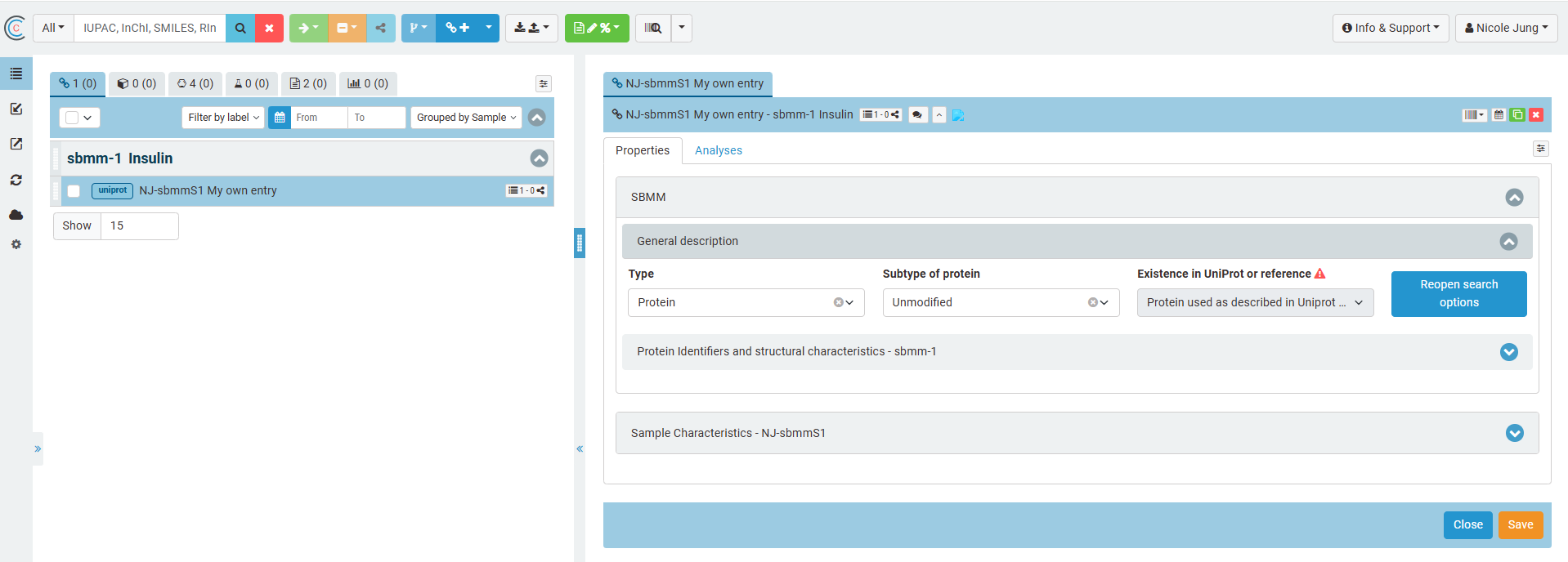
If a UniProt reference is available, the user can search for the UniProt entry by UniProT AC number or name and can select amongst the available UniProt resources. The selected Uniprot entry can be used to extract all relevant information on the protein's structure and characteristics. The information is added to the Chemotion SBMM ans summarized in the section "protein identifiers and structual characteristics". The extracted information is assigned to the SBMM concept related to the unique sequence of the protein and cannot be changed.

If a protein is modified, that means that a UniProt entry is used as a basis and the sequence is changed in an experiment or to conduct an experiment, the user can select the option "used modified protein". Along with the section "protein identifiers and structual characteristics of reference entries" another section "properties of the modified sequence or own protein" appears and can be used to define the modifications. The system will keep the sequence of the modified SBMM as identifier for the new SBMM concept and references the original entry in UniProt.

Adding information to the SBMM sample
If the user applies the SBMM in a reaction or wants to assign analytical data to it, the SBMM sample is relevant as all batch relevant information is stored in the sample. The sample information is the second part of the UI representation of an SBMM and is defined in the section "Sample Characteristics".
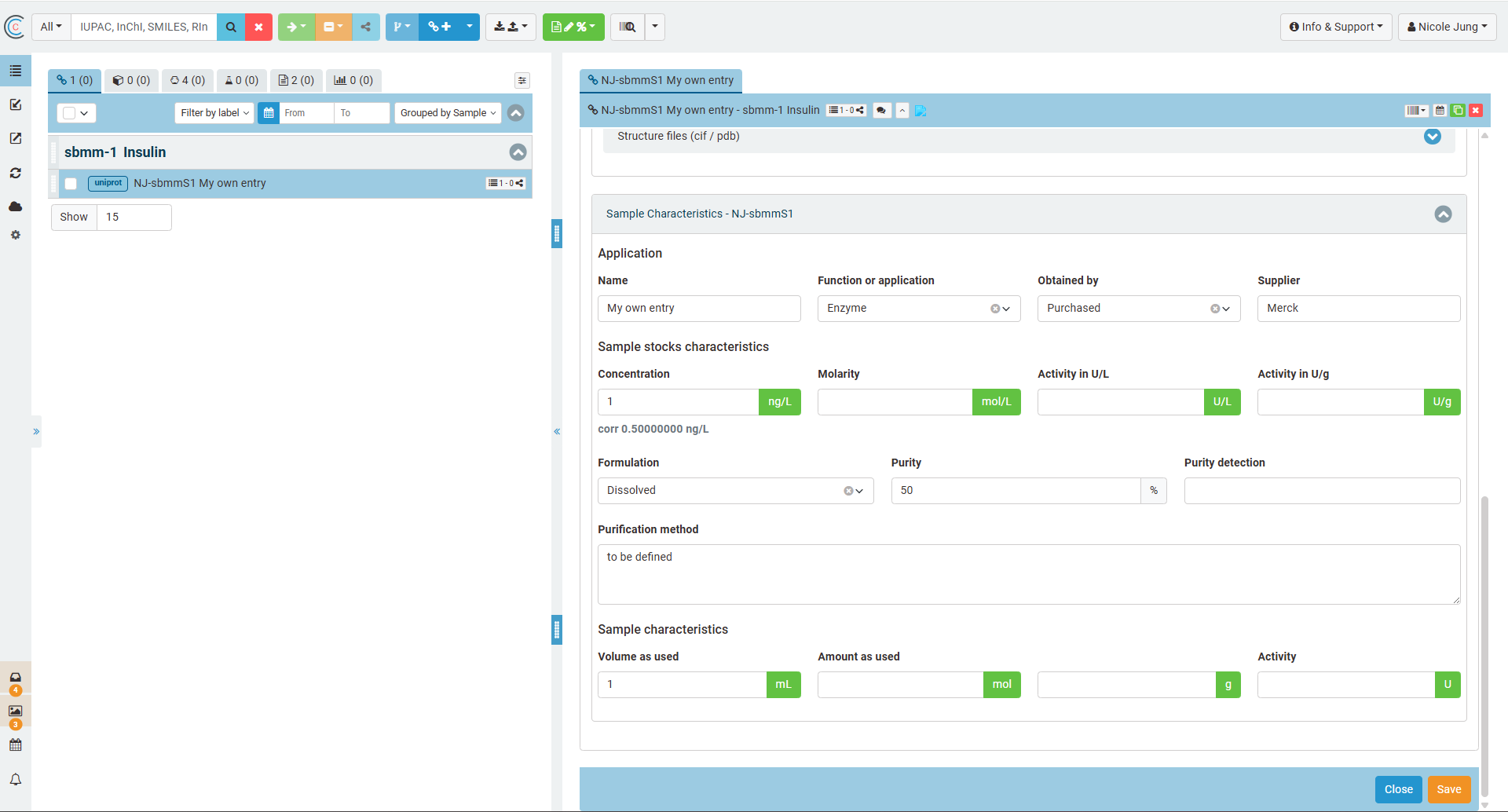
The following screenshot highlights again the differences of SBMM concept (blue) and SBMM sample (orange) and the way how to identify both in the list of elements.
